OUR APPROACHES, TOOLS, TECHNOLOGY:
Our general principle is to observe proceses in action, measure parameters quantitatively and complement approaches like mathematical modeling, molecular simulations etc. We strive to employ the most appropriate technique that allows us the best possible quantitative measurements for a given sample/ experimental system. Brief descriptions of major techniques used in the lab and some example movies from past publications:
Lattice Light-Sheet Microscopy:
The ability of this technique to acquire volumetric images of whole cells at high temporal resolution allows us to investigate endosomal dynamics, dorsal dynamic membrane topological changes, etc. Examples of movies from on-going investigations can be found under Research – endosomal trafficking.
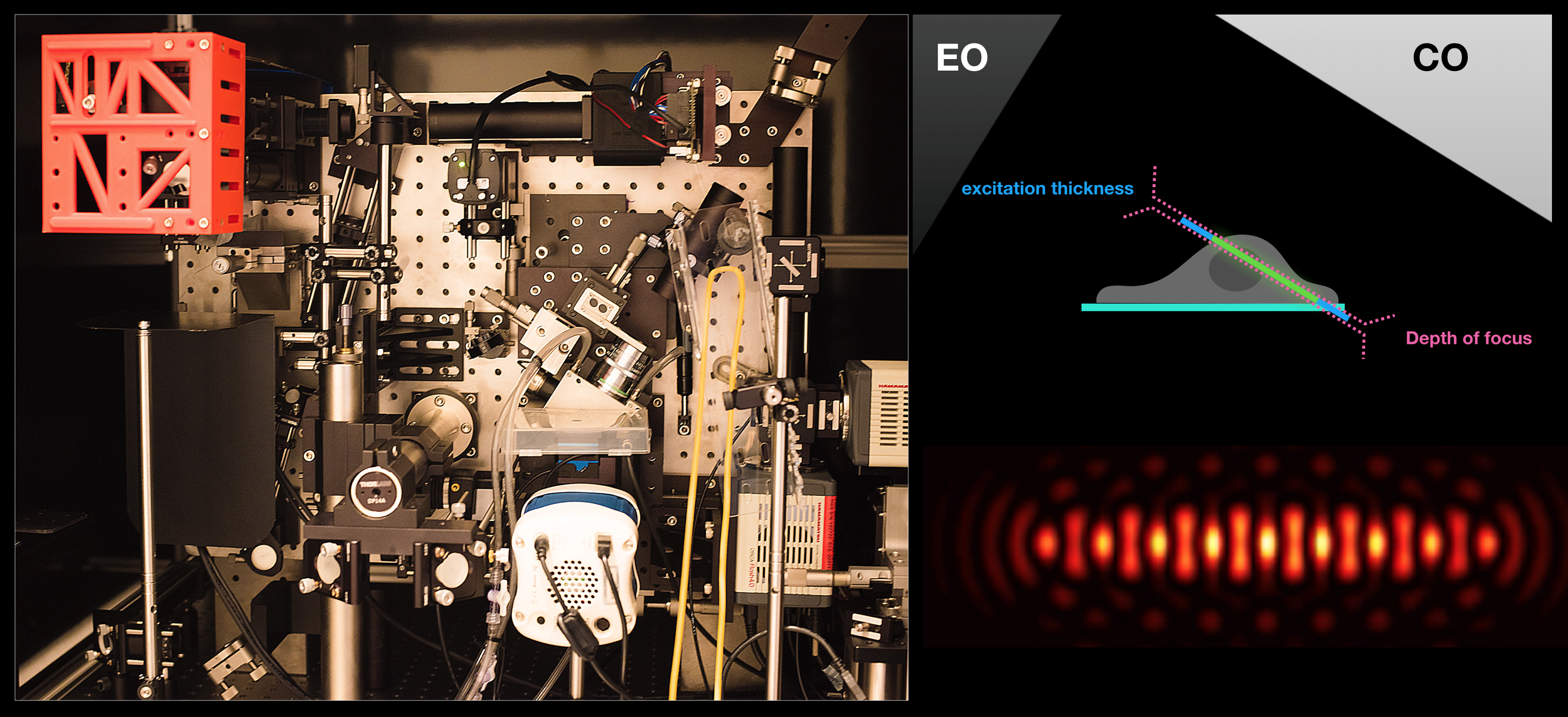
Optogenetics + LLSM:
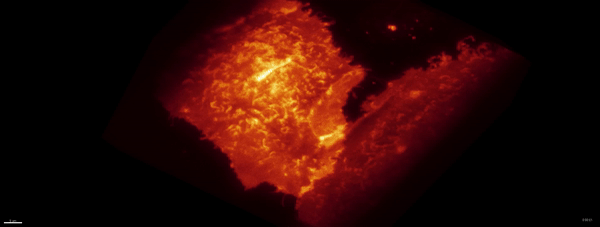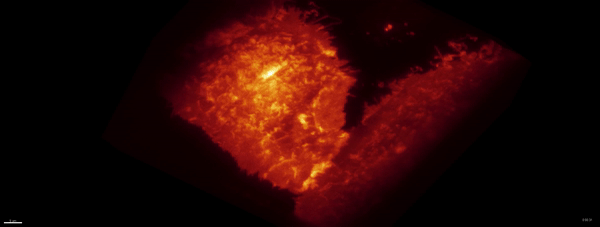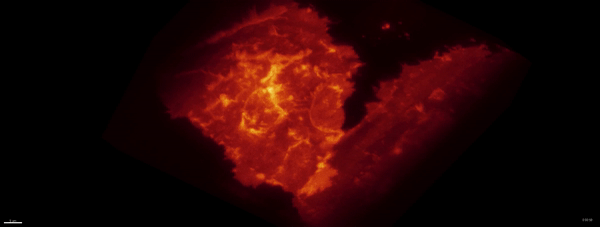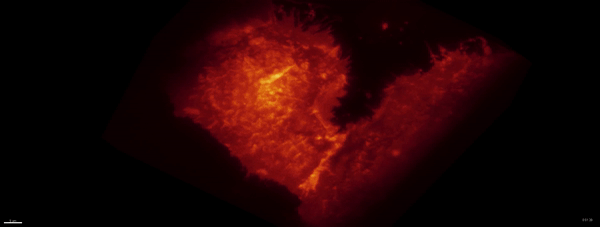
Movie:
PA Rac1 activated under LLSM.
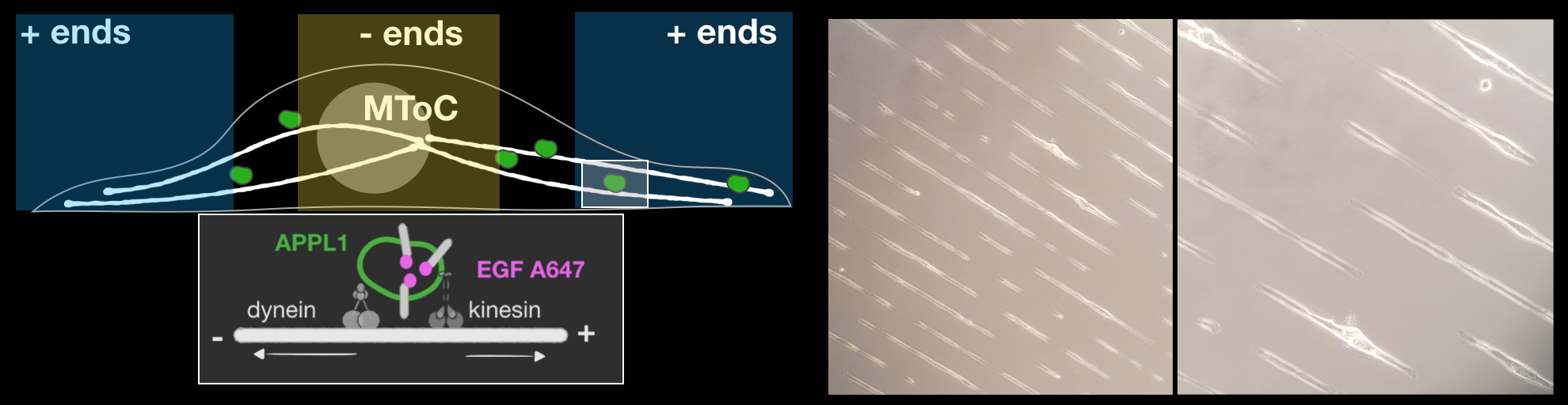
Micropatterning:
Micropatterning allows manipulation of the ‘foot-print’ of the cells on coverslips. This allows us to measure directionalities of endosomal motilities more robustly. (York et al. BioRxiv, 2018)
Analysis routines:
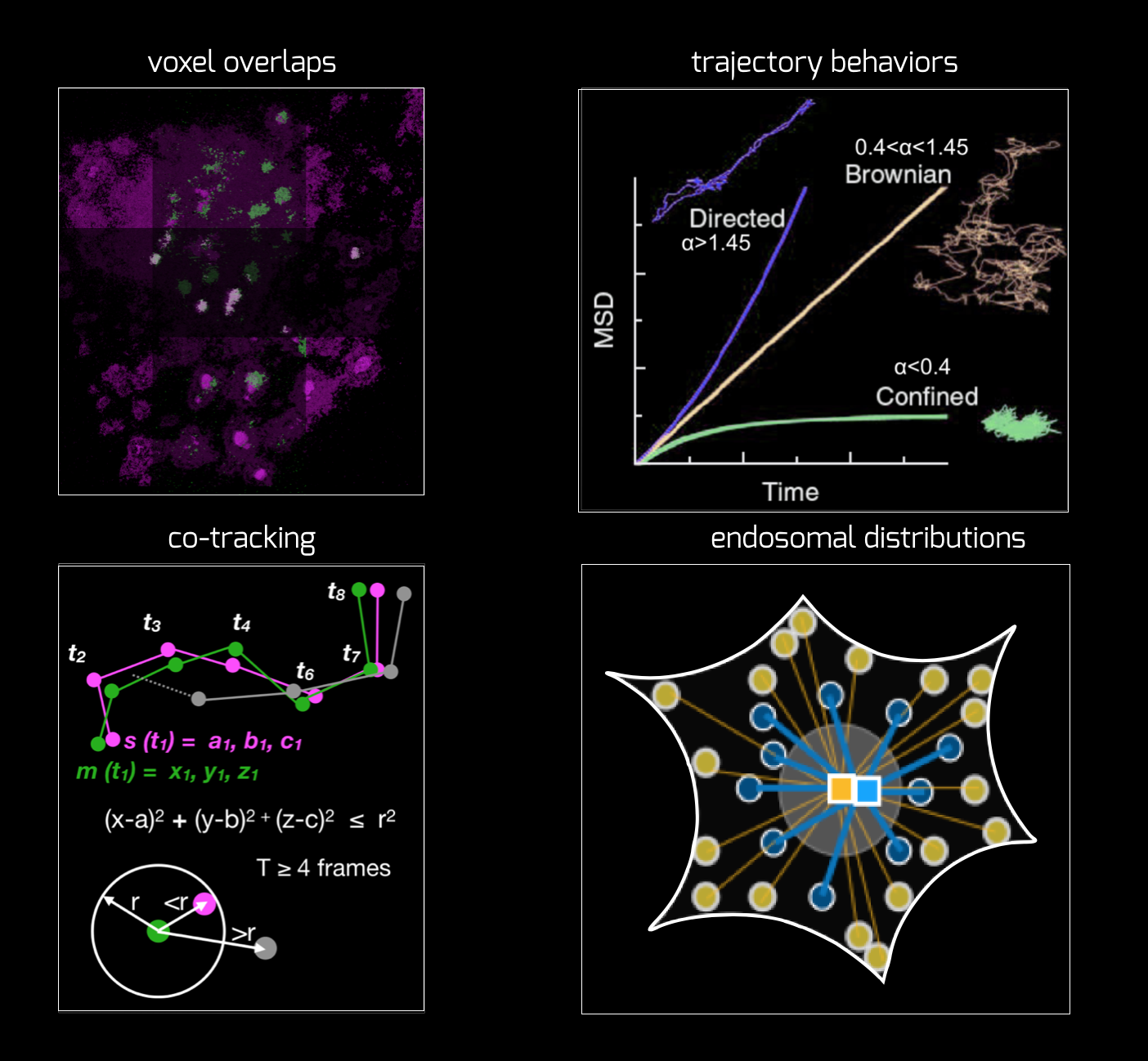
We develop some in-house methods as well as use a lot of codes from elsewhere. We strive to make it broadly usable and open-source. We collaborate with Tomasz Bednarz from UNSW arts and Srigokul Upadhyayula (Harvard) on this front. Currently, we are working on developing a cross-channel intensity as well as mobility analyser, that will allow coorelating mobility with various signals from various endosomal markers.
Single Molecule approaches:
We utilise single molecule methodologies based on imaging (TIRF) or Fluorescence Correlation Spectroscopy (FCS) depending on the experimental parameters to be measured. With our previous experiences, we are well-positioned to combine this with other methodologies as required. (Arumugam et al. PNAS, 2014)
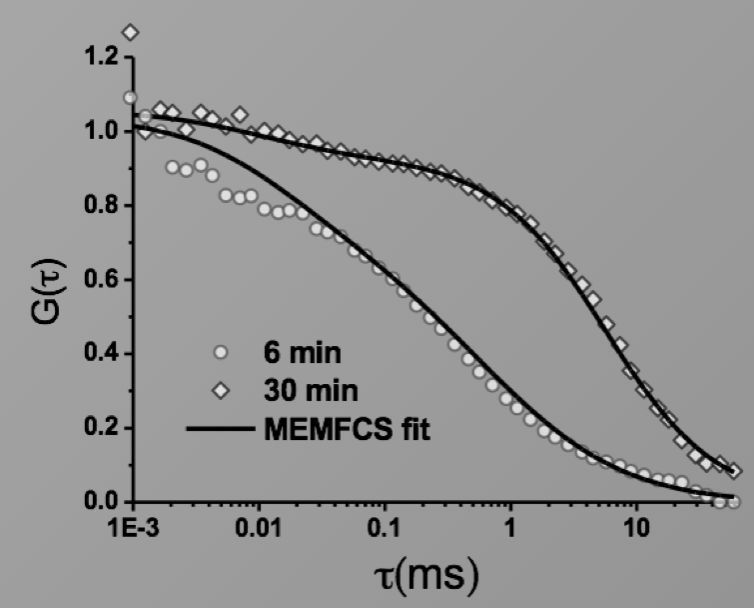
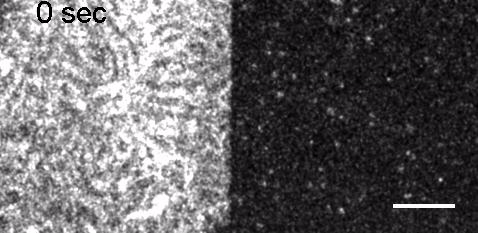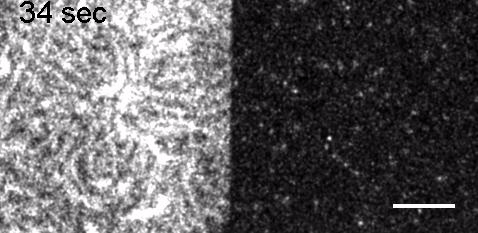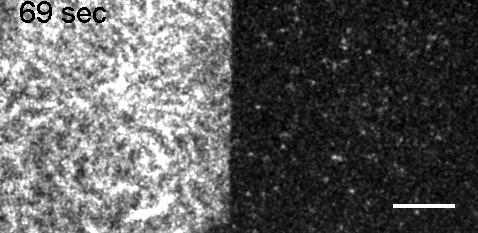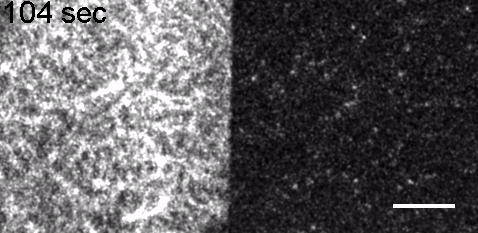
Image: FCS of StxB clustering on membranes. Movie:TIRF imaging of bulk FtsZ -YFP(left) and 20 nM FtsZ-A647 (right).
Fluorescence Recovery After Photobleaching (FRAP) and Fluorescence Loss in Photobleaching (FLIP):
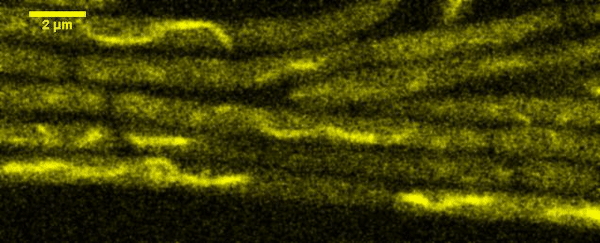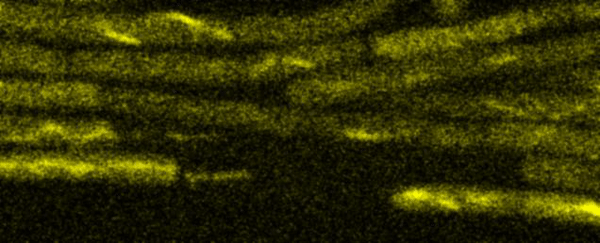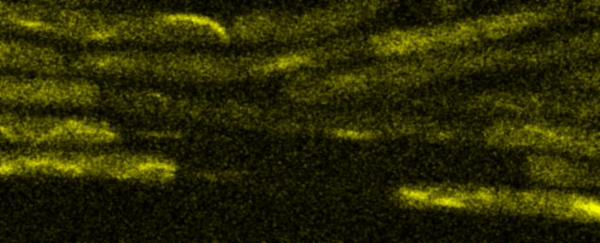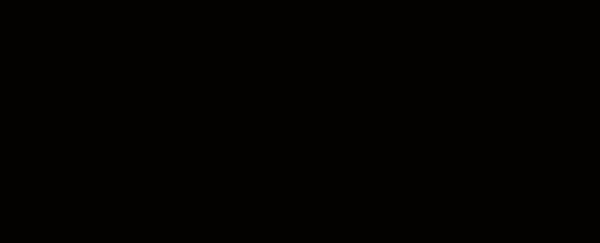
Artificial membrane systems:
Supported lipid bilayer based systems to study complex protein membrane interactions:
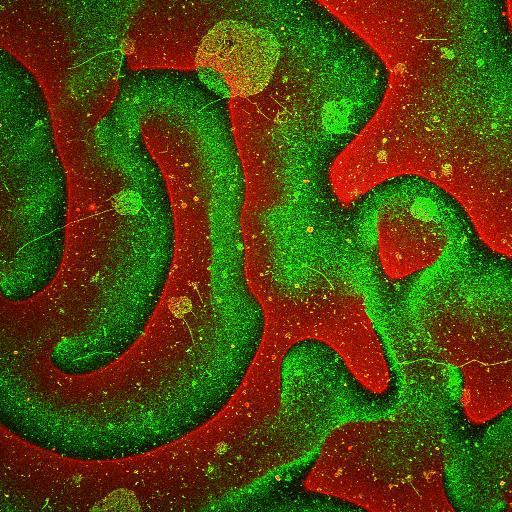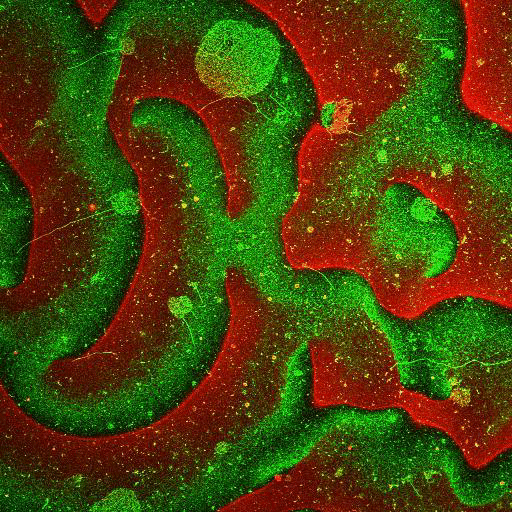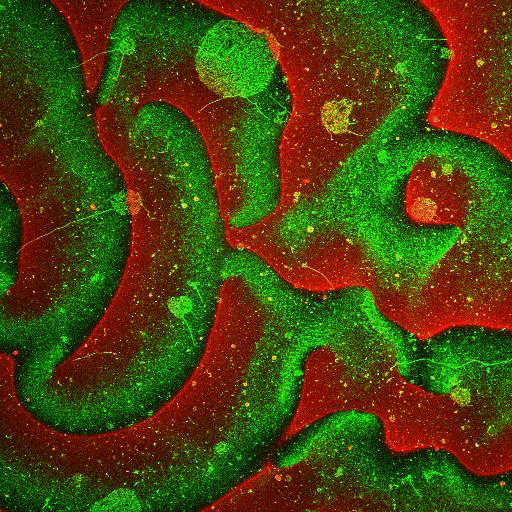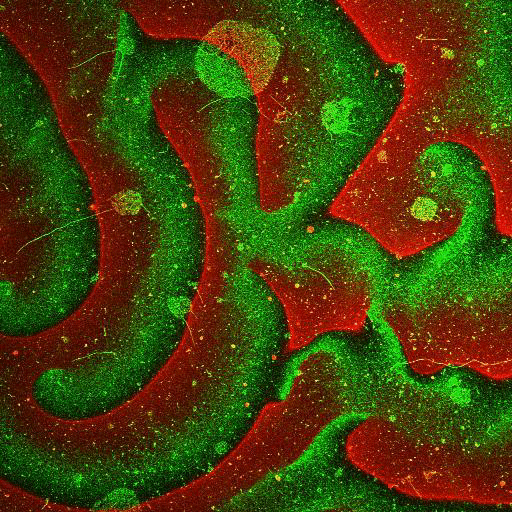
Movie: Pattern formation by Min CDE + FtsZ on a supported lipid bilayer in the presence of ATP and GTP. The pattern formation is a result of reaction-diffusion based systems which, in this case, require membranes as a substrate to allow slow–diffusion of MinD. (Arumugam et al. PNAS, 2014)
Giant Unilamellar based systems for studying phase separation, membrane-cytoskeleton interactions etc.:
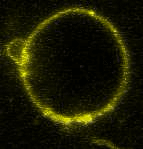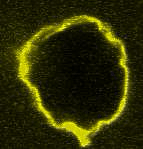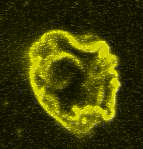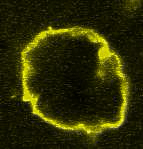
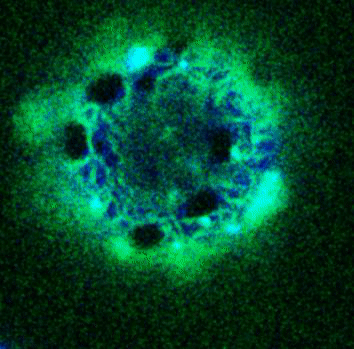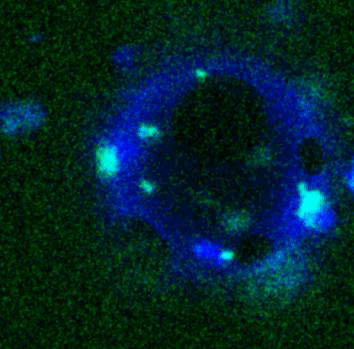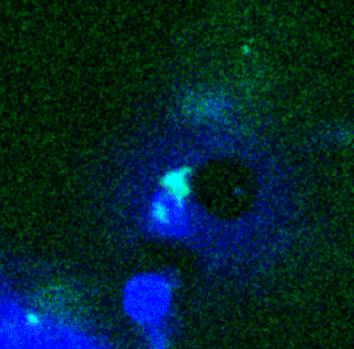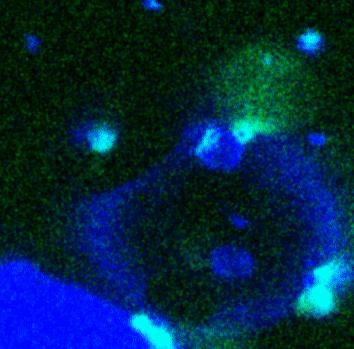
Movie:
FtsZ coated vesicle under osmotic shocks.(Arumugam et al. Ang. Chemie. 2012)
Disassembly of FtsZ filaments on a phase separated GUVs by MinC.(Arumugam et al. Biophys. J. 2015)
